My name is Marcel Schilling and I’m a Senior Bioinformatician.
My research focusses on post-transcriptional regulation in the context of Alzheimer’s Disease.

My research focusses on post-transcriptional regulation in the context of Alzheimer’s Disease.


I studied Bioinformatics (BSc & MSc) at Freie Universität Berlin (FU Berlin) working as a research assistant at the Max Planck Institute for Molecular Genetics (MPI MolGen). There, I computationally predicted the effects of single nucleotide polymorphisms on microRNA-targeting in the context of neuropsychiatric diseases.
After completing my MSc studies, I joined the N. Rajewsky lab at the Berlin Institute for Medical Systems Biology (BIMSB) / MDC. There, I first worked on a project developing a method to recover miRNA-target interactions from sRNA-seq data based on chimeric reads before gaining experience in the developing field of circular RNAs. For my main project, I developed a 3-dimensional physical model of the C. elegans gonad and a computational pipeline to map spatially resolved RNA-seq data onto it. We used the resulting virtual in-situ hybridization assays to investigate post-transcriptional regulation throughout the germline development from stem cells to mature oocytes.
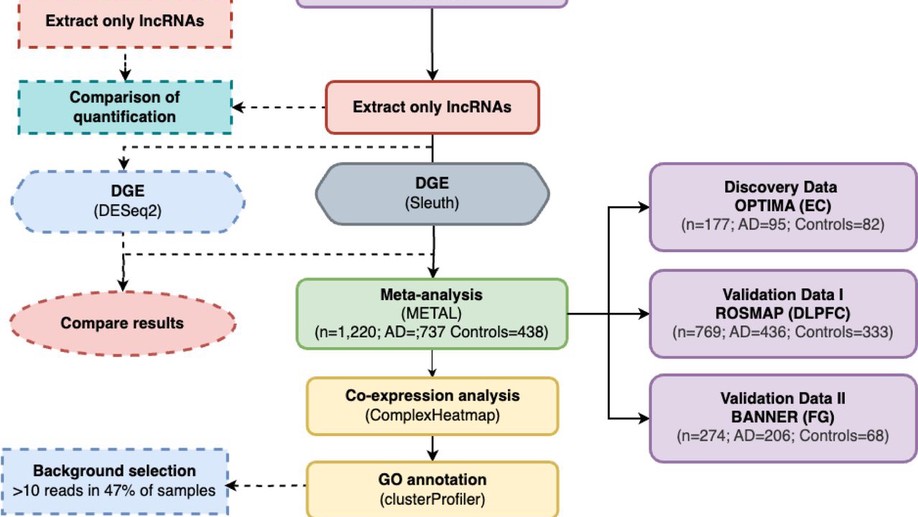
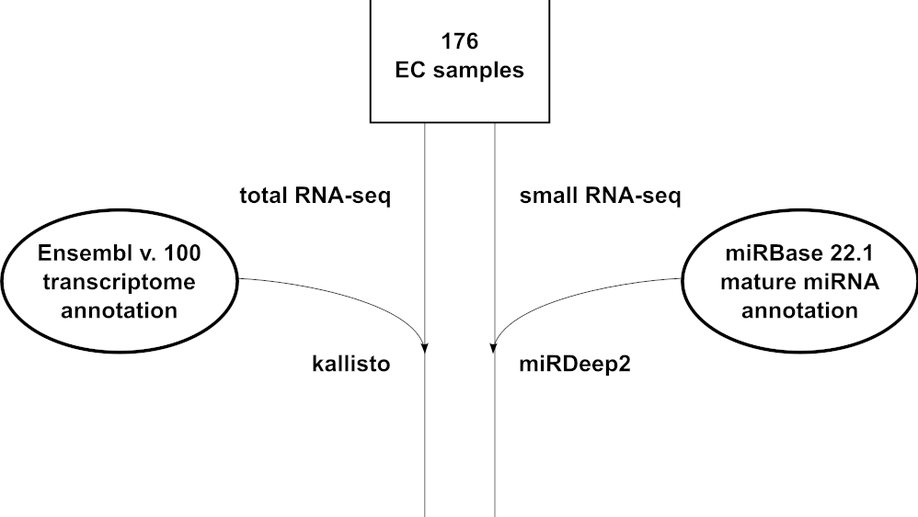
Next, I investigated differential expression in Alzheimer’s patients brains at the Lübeck Interdisciplinary Platform for Genome Analytics (LIGA) at Universität zu Lübeck (UzL) before joining the Gene Regulation of Cell Identity group at P-CMR[C]/IDIBELL, where I continued my research on post-transcriptional regulation in the context of early Alzheimer’s Disease using single-cell RNA-seq techniques on an in-vitro human induced pluripotent stem cell model.
Since, I joined the Francesc Cebrià Lab at University of Barcelona (UB) where I am analysing data of planarians.
MSc in Bioinformatics, 2011 - 2013
Freie Universität Berlin
BSc in Bioinformatics, 2008 - 2011
Freie Universität Berlin
These are the languages I like to code in
90%
70%
95%
50%
Software that helps me organize my pipelines
85%
75%
90%
60%
MY ONGOING QUEST TO LEARN, EXPLORE, INVESTIGATE, AND SHARE WHAT I HAVE LEARNED
Courses I completed and honors I received
For a full list, see my ORCiD
I really enjoy exchange about research