Abstract
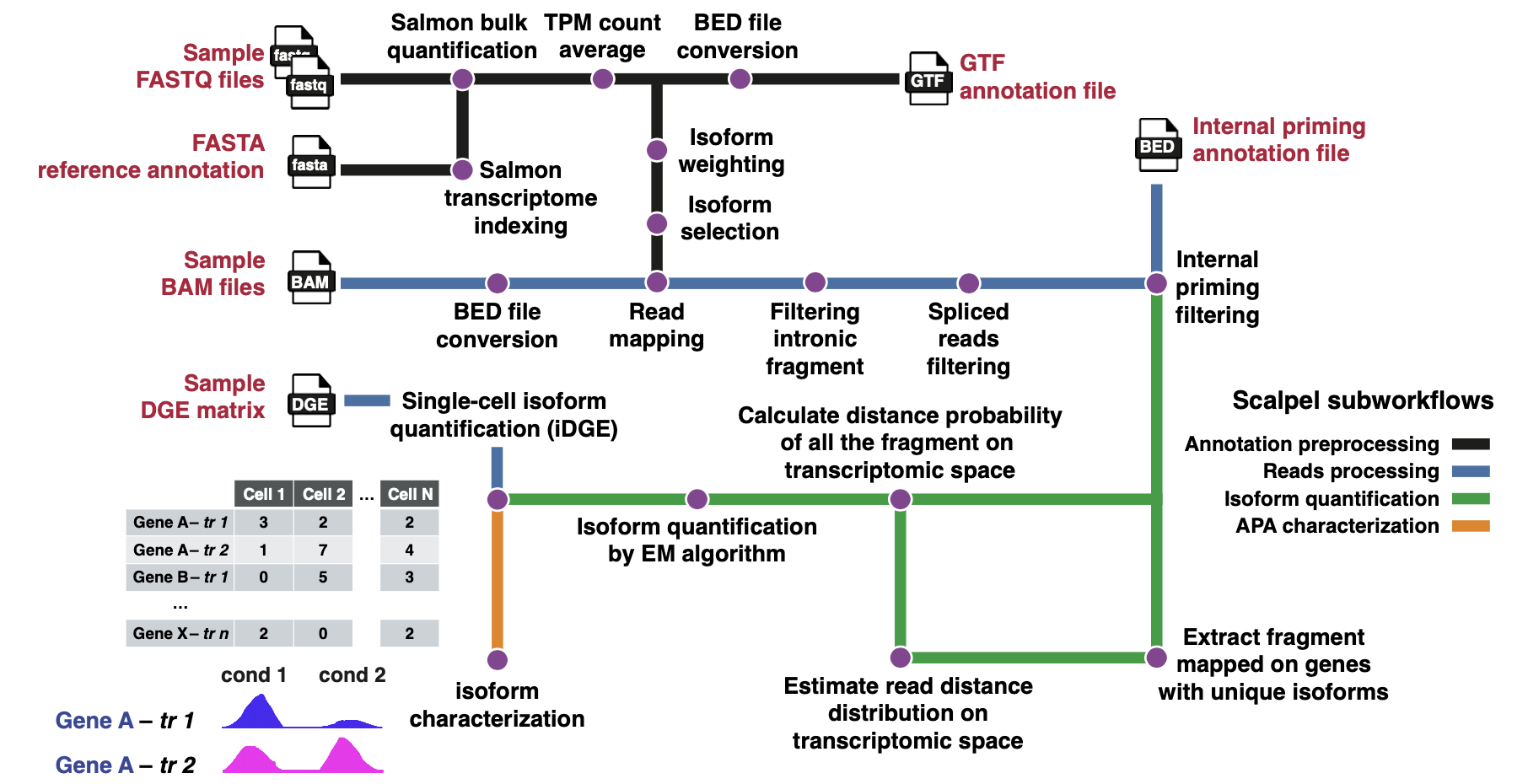
Single-cell RNA sequencing (scRNA-seq) has facilitated the study of gene expression and the development of new tools to quantify transcript in individual cells. Yet, most of these methods have low sensitivity and accuracy. Here we present SCALPEL, a Nextflow-based tool to quantify and characterize transcript isoforms at the single-cell level using standard 3’ based scRNA-seq data. SCALPEL predictions have higher sensitivity than other tools and can be validated experimentally. We have used SCALPEL to study the changes in isoform usage during mouse spermatogenesis and in the differentiation of induced pluripotent stem cells (iPSCs) to neural progenitors. These analyses allow the identification of novel cell populations that cannot be defined using conventional gene expression profiles, confirm known changes in 3’ UTR length during cell differentiation, and identify cell-type specific miRNA signatures controlling isoform expression in individual cells. Together, our work highlights how SCALPEL expands the current scRNA-seq toolset to explore post-transcriptional gene regulation in individual cells from different species, tissues, and technologies to investigate the variability and the specificity of gene regulatory mechanisms at the single-cell level.