Abstract
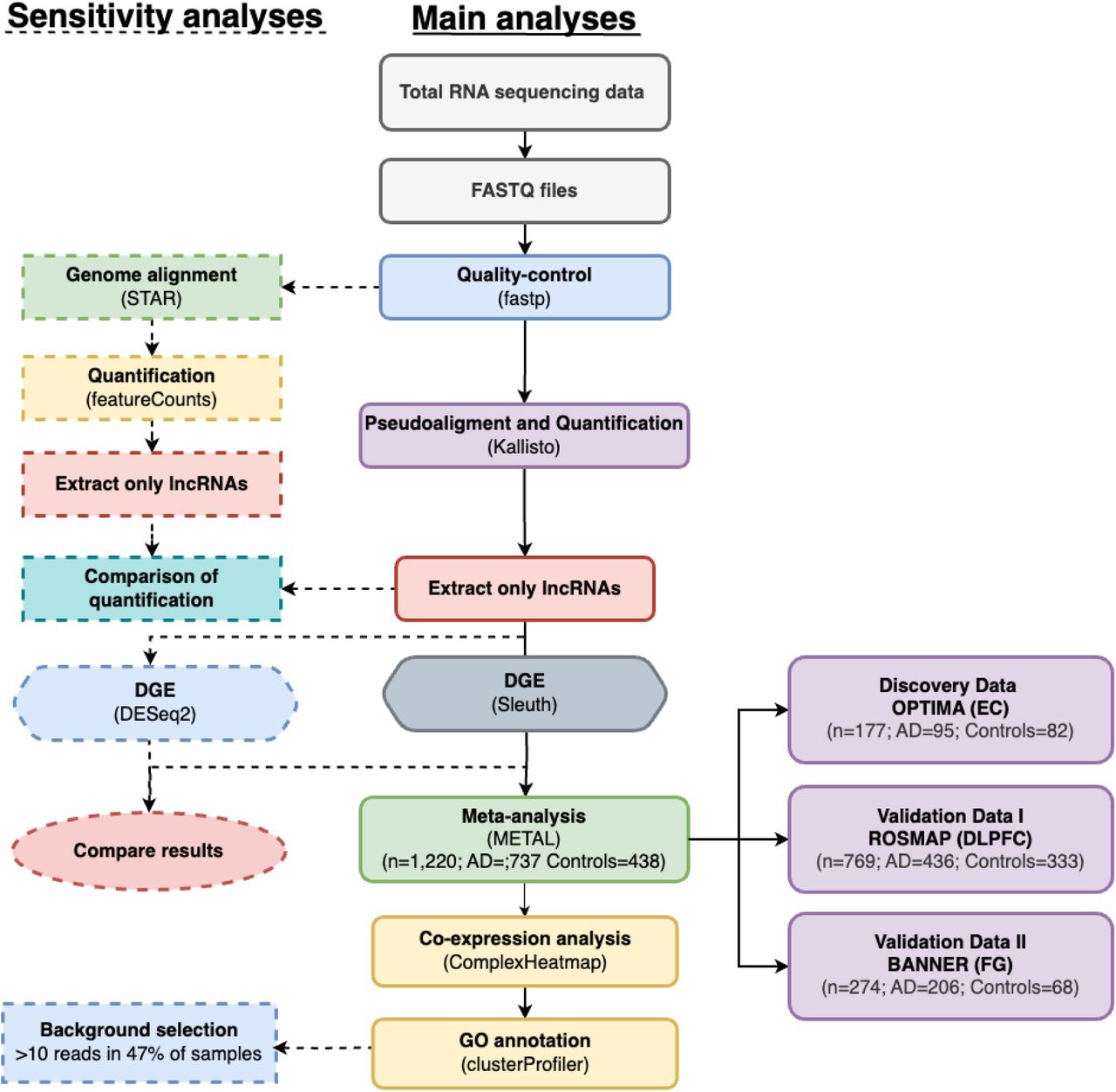
Long non-coding RNAs (lncRNAs) have been reported to show differential expression in Alzheimer’s disease (AD), albeit with inconsistent results. In addition, only a few studies explored lncRNA expression in human brain samples. In this study, we performed differential gene expression (DGE) analyses on lncRNA expression data (derived from RNA sequencing [RNA-seq]) from a total of n=1,220 samples collected from three different human brain regions. These included 177 individuals drawn from the OPTIMA study (discovery data; n[AD cases] = 95, n[controls] = 82) collected from entorhinal cortex (EC). In addition, we analyzed independent RNA-seq data from the ROSMAP (dorsolateral prefrontal cortex; n[AD cases] = 436, n[controls] = 333]) and BANNER studies (fusiform gyrus; n[AD cases] = 206, n[controls] = 68). In the discovery dataset (EC), we identified 121 significantly differentially expressed lncRNAs after FDR correction. Meta-analysis with ROSMAP and BANNER data resulted in a total of 356 significantly differentially expressed lncRNAs, showing the same direction of effect in all three datasets. Besides replicating many AD-related lncRNAs, we also identified several dozen lncRNAs not previously linked to AD, including Lnc-TNFRSF13B-2, CSRP1-AS1, Lnc-MIB2-1, Lnc-TTC5-2, and GSN-AS1. GO annotation highlighted numerous relevant processes, including neuron development, synapse function, and axon development. To our knowledge, our study comprises the largest RNA-seq-based DGE analysis of the human brain and is the first to utilize EC samples, the site where progressive AD pathology originates. With its large number of novel AD-associated lncRNAs it provides a vastly extended reference for future work on this topic.